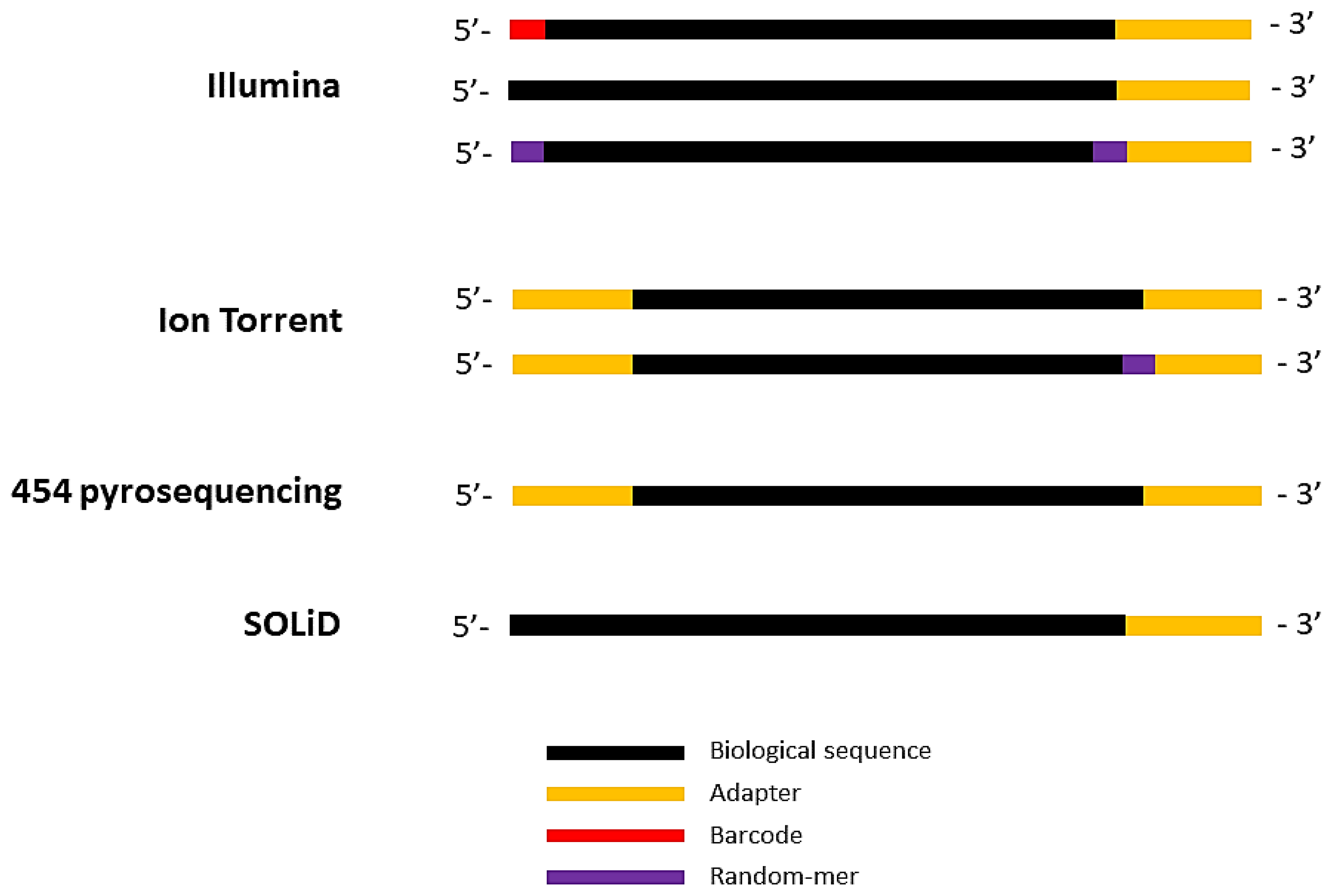
Biomolecules | Free Full-Text | High-Throughput Identification of Adapters in Single-Read Sequencing Data

PathoQC: Computationally Efficient Read Preprocessing and Quality Control for High-Throughput Sequencing Data Sets - Changjin Hong, Solaiappan Manimaran, William Evan Johnson, 2014
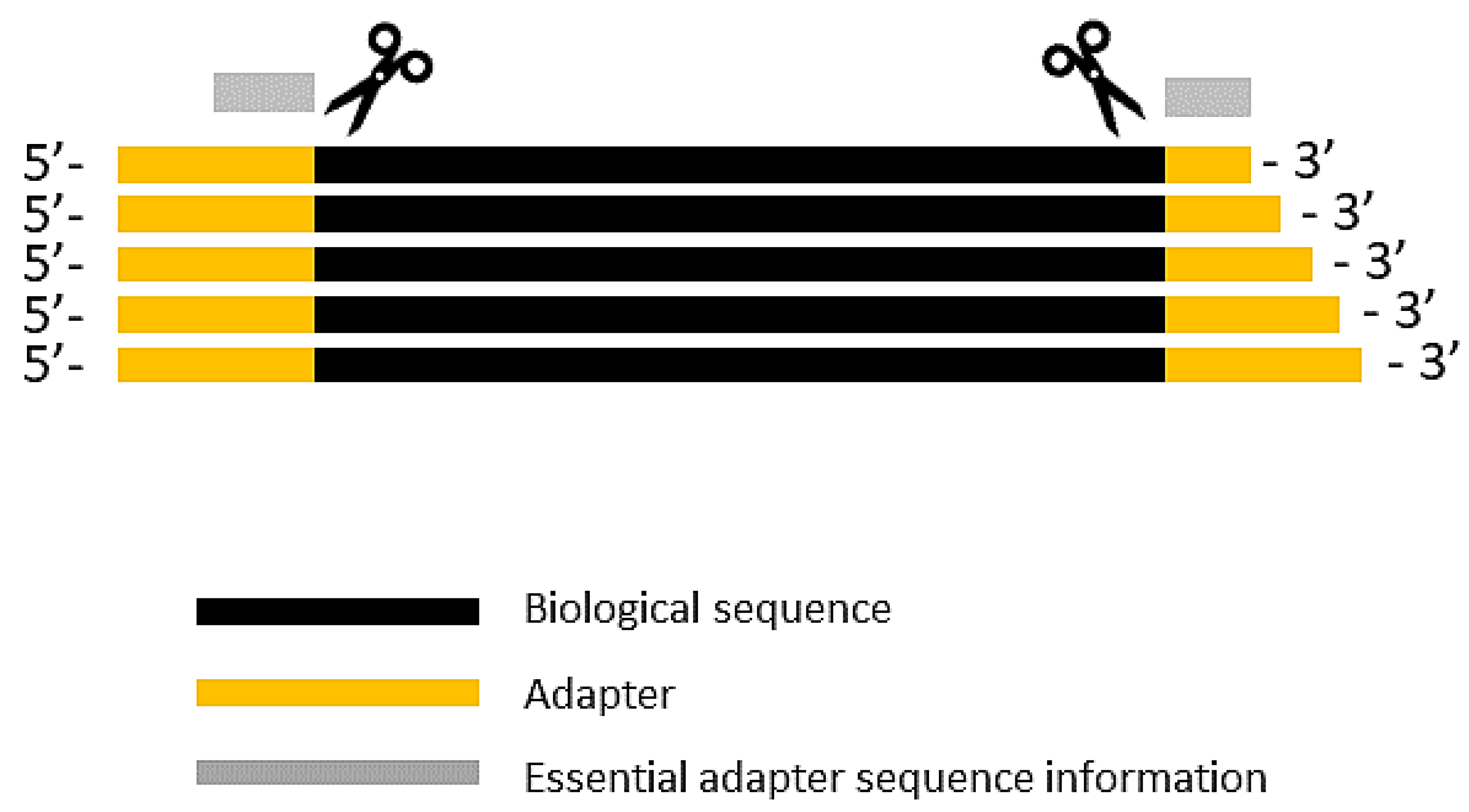
Biomolecules | Free Full-Text | High-Throughput Identification of Adapters in Single-Read Sequencing Data
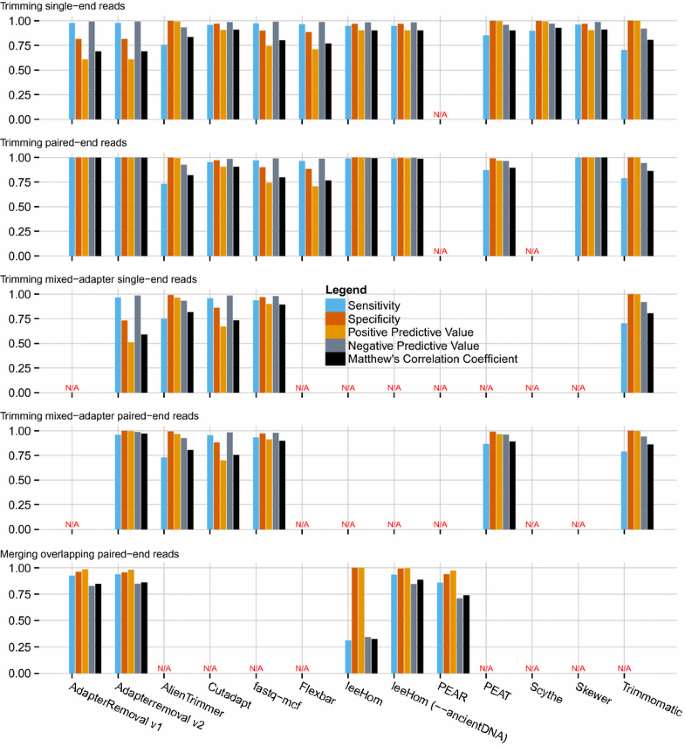
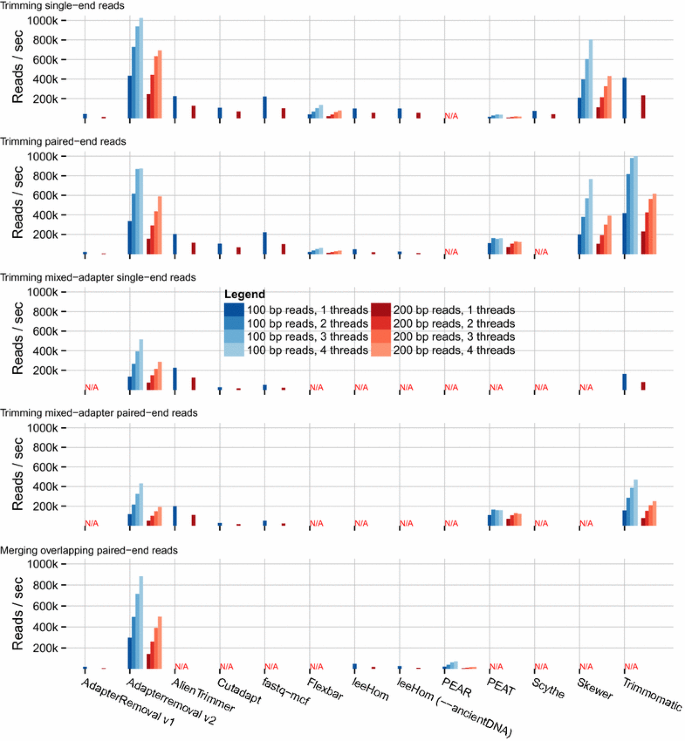
AdapterRemoval v2: rapid adapter trimming, identification, and read merging | BMC Research Notes | Full Text
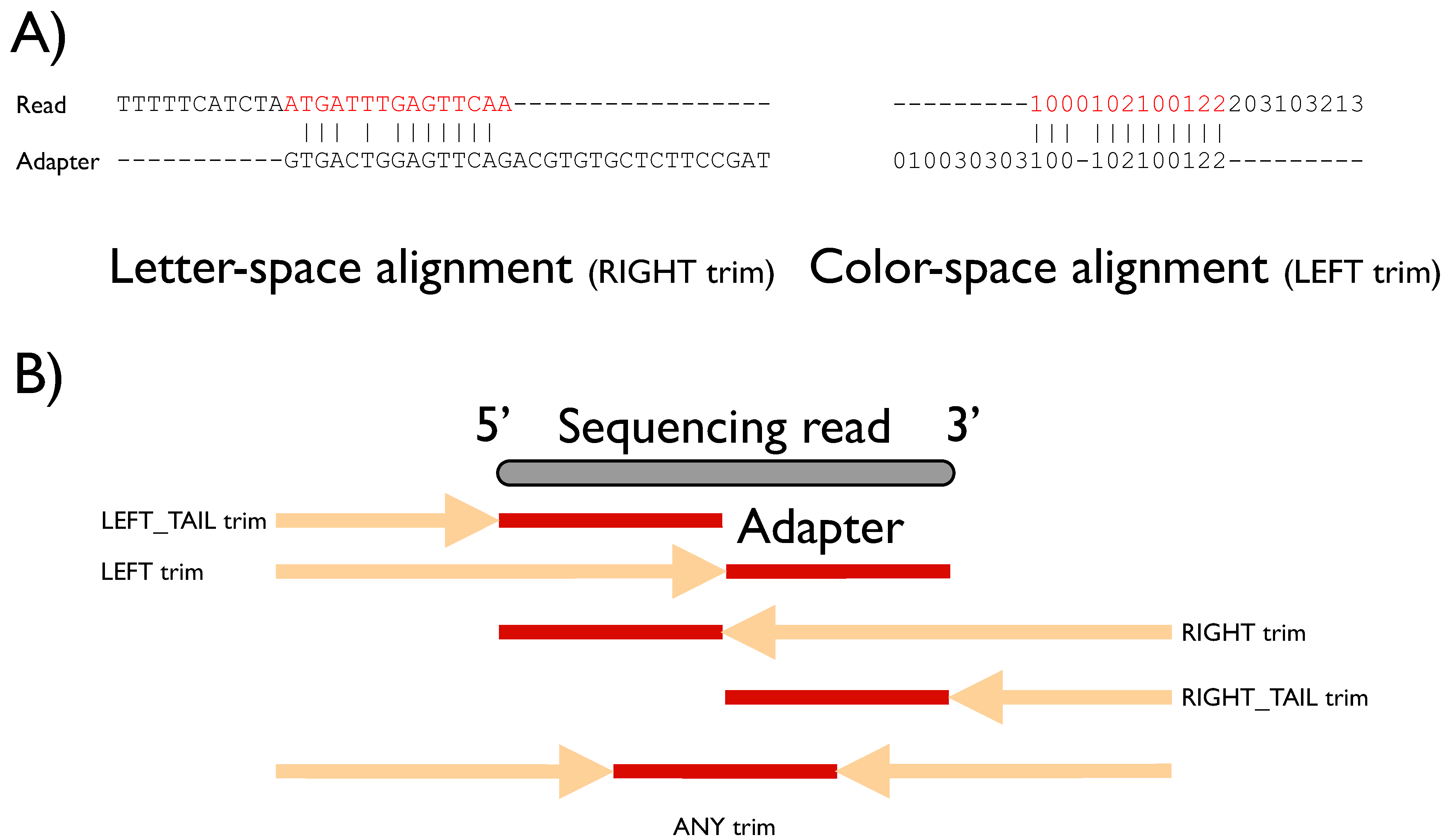
Biology | Free Full-Text | FLEXBAR—Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms

Kraken: A set of tools for quality control and analysis of high-throughput sequence data☆ | Semantic Scholar

Assessing the utility of long-read nanopore sequencing for rapid and efficient characterization of mobile element insertions - Laboratory Investigation
GitHub - naturalis/galaxy-tool-cutadapt-sequence-trimmer: Use cutadapt to trim and discard reads and read sections.